Biology with XFELs
XFELs are unique light sources that can be used to explore matter at atomic length and femtosecond time scales. The increase in brightness along with ultra-short pulses has facilitated the appearance of a new application of XFEL technology in the field of structural biology via serial femtosecond crystallography (SFX).
SFX enables unique experiments on many different types of biomolecules under nearly “native” conditions at room temperature. Below is a glimpse of some of the experiment that scientists can develop at XFELs.
1. Mapping an Important Drug Target: GPCRs
G protein-coupled receptors (GPCRs) are a large family of cell surface receptors involved in several of the most chronic human pathologies such as cancer, cardiovascular diseases, metabolic disorders (e.g., obesity and diabetes) and immune disorders and therefore are extremely attractive therapeutic targets. It has been estimated that one-third to one-half of all marketed drugs act by binding to GPCRs. SFX has rapidly accelerated structural studies of GPCRs in the past decade. Two examples of this success are shown below.
Unravelling Structure of the Angiotensin receptor: a Breakthrough in regulation of Blood Pressure
When angiotensin hormones bind to the angiotensin receptor of the corresponding G-protein, it signals the constriction of blood vessels which leads to higher blood pressures. Blocking the hormones’ binding with a drug can lead to reduced blood pressures. Unfortunately, blocking this receptor also blocks the other, typically beneficial, actions of the G-protein coupled receptor, such as preventing dizziness. Thus, understanding of drug targets for hypertension implies elucidating the binding pocket of the seemingly similar angiotensin receptors AT1R and AT2R. The similar pockets of these two angiotensin receptors imply that similar drugs would be effective; however, this was not the case in clinical studies, drugs that were successful to treat issues related to AT1R were not nearly as successful as treating those related to AT2R. The 3D structure of AT1R obtained at XFELs showed an unexpected result: although they bound to the same pocket, they bound much differently, leaving the site in an active state but inhibiting the binding of the G-protein, thus rendering it overall inhibited. This finding demonstrates the power of structure-based drug discovery with XFELs. Now drugs can be designed from the ground up designed to match the pocket, rather than relying on educated guesses of similar pockets.
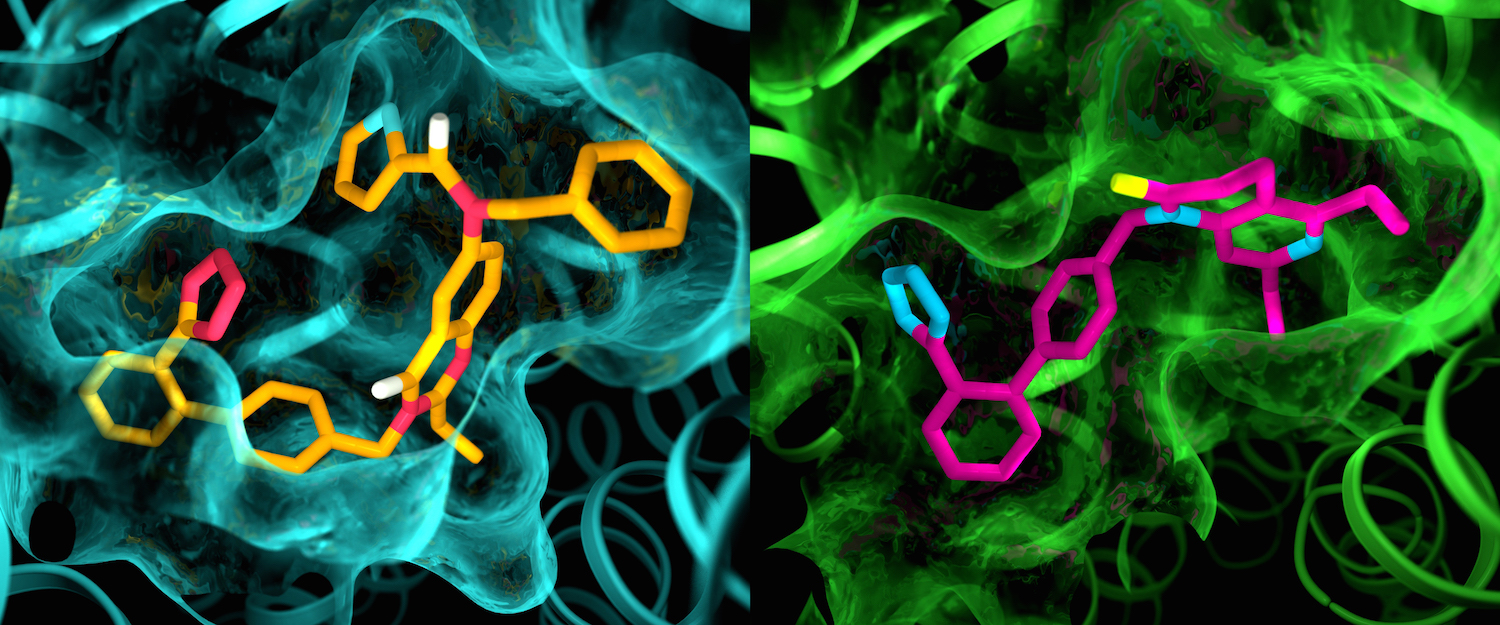
Although the two angiotensin II receptors are thought to be very similar, an X-ray study showed clear differences in the pockets where the receptors bind to drug-like compounds. This illustration shows details in the pocket structures of AT2 (left) and AT1 (right). Image by Greg Stewart/SLAC National Accelerator Laboratory.
3D structures of Melatonin-2 receptor for advancing in the development of drugs for sleep disorders
The human MT1 and MT2 melatonin receptors are GPCRs that help to regulate circadian rhythm and sleep patterns. Drug development efforts have targeted both receptors for the treatment of insomnia, circadian rhythm and mood disorders, and cancer, and MT2 has also been implicated in type 2 diabetes. The 3D structures of the human MT2 receptor reported at XFELs, and their comparison with the human MT1 receptor reveal that, despite conservation of the orthosteric ligand-binding site residues, there are notable conformational variations as well as differences in melatonin dissociation kinetics that provide insights into the selectivity between melatonin receptor subtypes. Although, a membrane-buried lateral ligand entry channel is observed in both proteins, the MT2 structures reveal a narrow opening towards the solvent in the extracellular part of the receptor. This finding contributes to a molecular understanding of melatonin receptor subtype selectivity and ligand access modes, which are essential for the design of highly selective melatonin tool compounds and therapeutic agents.
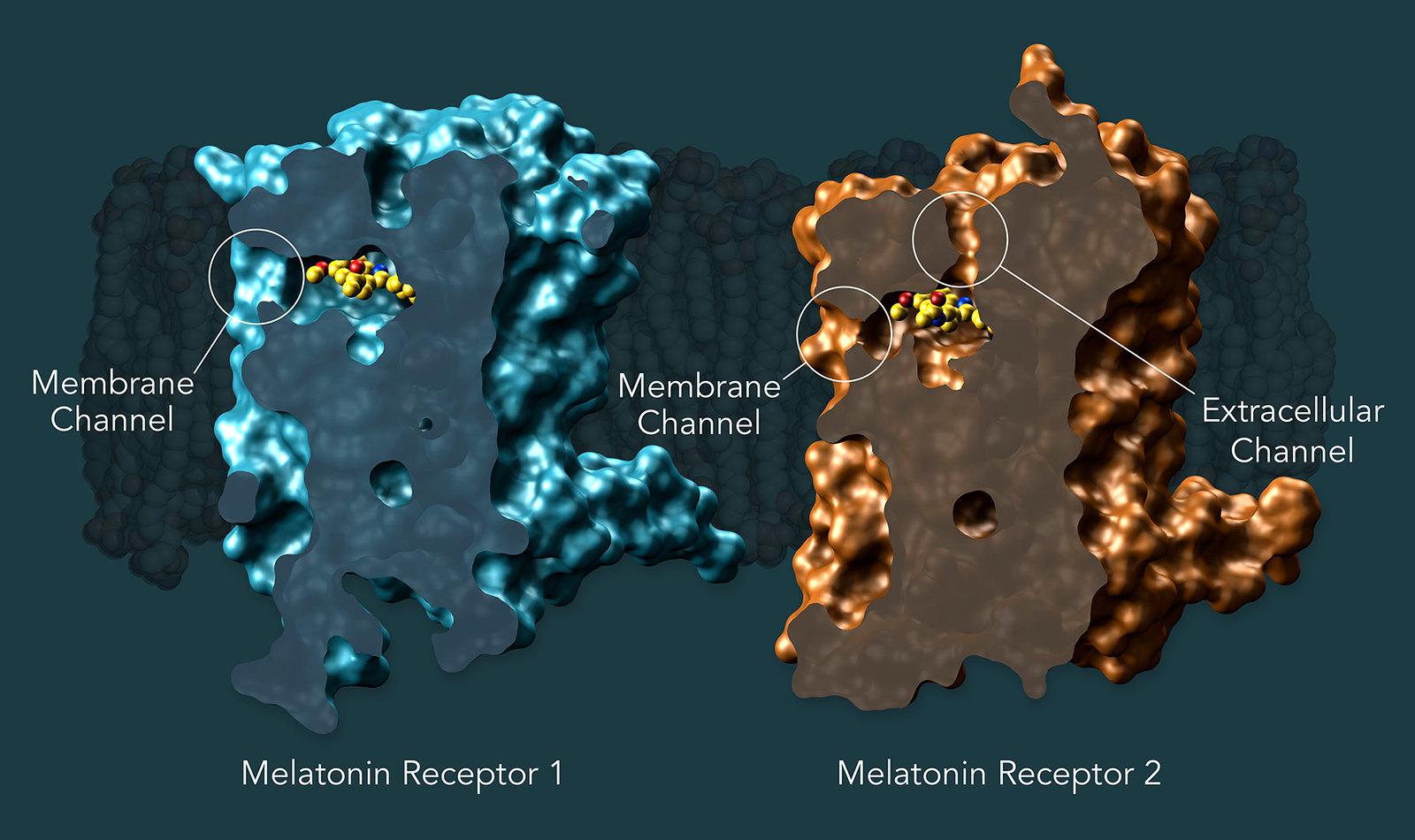
Researchers showed that both melatonin receptors contain narrow channels embedded in the cell’s fatty membranes. These channels only allow melatonin, which can exist happily in both water and fat, to pass through, preventing serotonin, which has a similar structure but is only happy in watery environments, from binding to the receptor. They also uncovered how some much larger compounds only target MT1 despite the structural similarities between the two receptors. Image by Greg Stewart/SLAC National Accelerator Laboratory
2. Shining light on sleeping sickness
African trypanosomiasis disease, also called sleeping sickness, is a fatal disease caused by the protozoan parasite Trypanosoma brucei (Tb). People can get this parasite when an infected Tsetse fly bites them. Symptoms include fatigue, high fever, headaches, and muscle aches. If the disease is not treated, it can cause death. Two protein targets have been identified as promising drug targets to develop new treatments against sleeping sickness. The Trypanosoma brucei cysteine protease cathepsin B (TbCatB), which is involved in host protein degradation, and the inosine-5’-monophosphate dehydrogenase (IMPDH), a protein involved in maintaining the balance between guanylate deoxynucleotide and ribonucleotide levels that is pivotal for the parasite. The structure of the mature, active form of TbCatB and TbIMPDH have so far not provided sufficient information for the design of a safe and specific drug against T. brucei. By combining two recent innovations, in vivo crystallization and SFX with XFELs, the crystal structures of these two proteins have been reported to high-resolution. In the case of TbCatB the structure reveals the mechanism of native TbCatB inhibition. As for IMPDH, the structure reveals the presence of ATP and GMP at the canonical sites of the Bateman domains, the latter in a so far unknown coordination mode.
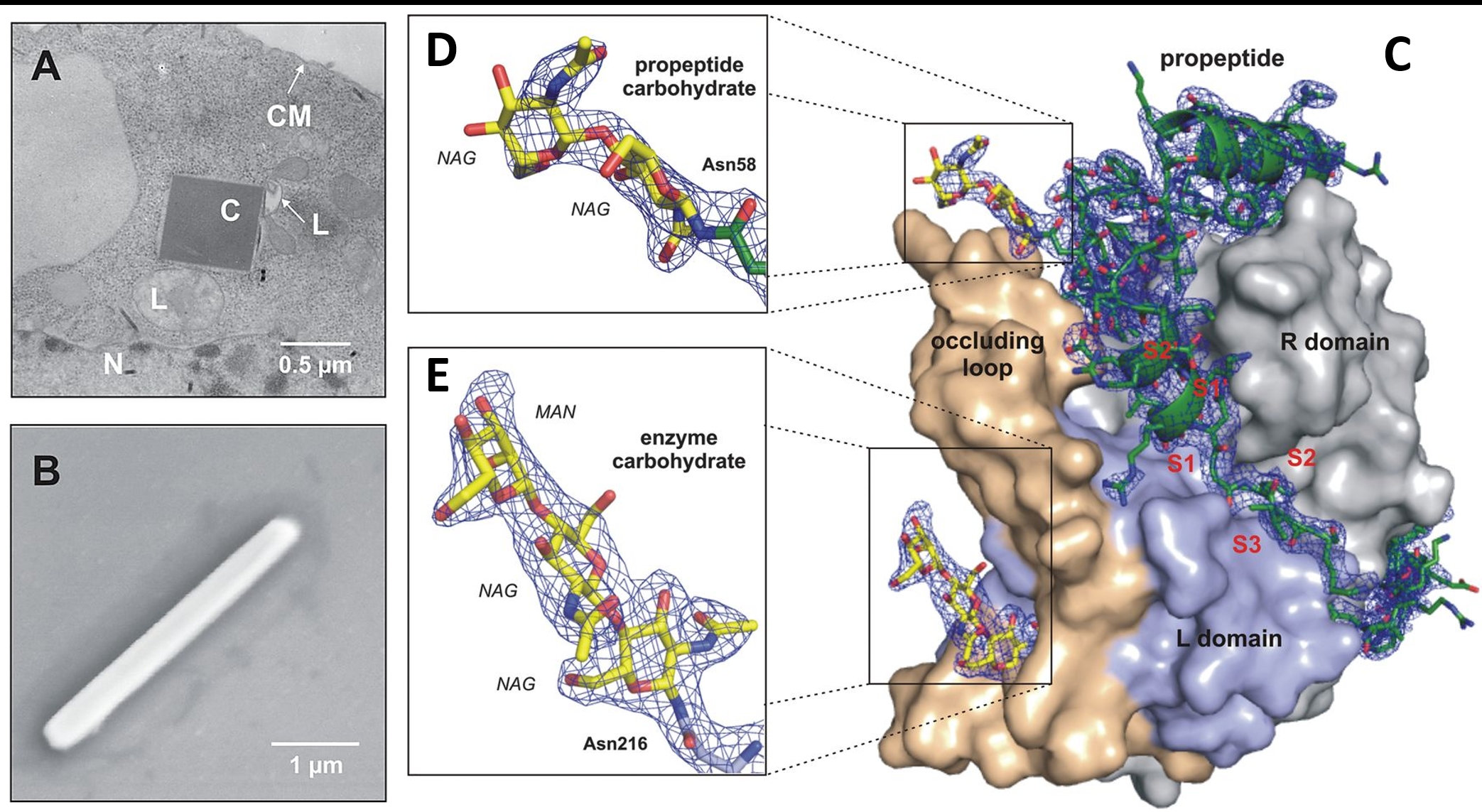
A) Transmission electron microscopy of an infected Sf9 insect cell showing a crystal of overexpressed TbCatB. B) Scanning EM of a single TbCatB crystal after isolation. C) Surface representation of the TbCatB-propeptide complex. The solution revealed additional electron density of the propeptide (green) that is bound to the V-shaped substrate-binding cleft and of two carbohydrate structures (yellow) N-linked to the propeptide (D) and to the mature enzyme (E).
3. Molecular Movie of the Trans/Cis Isomerization in Photoactive Yellow Protein
PYP is a small, soluble protein found in purple sulfur bacteria, where it senses blue light. The photocycle of PYP involves many separate, sequential steps that begin very quickly in femtoseconds, and end in about a second. A light-absorbing chromophore in the protein absorbs blue light and switches from a nearly straight (trans) conformation to a bent (cis) conformation in less than a picosecond. This conformational change is then thought to activate other sensing proteins in the bacterium, which control the direction that the bacterium is swimming. Finally, the chromophore shifts back to the straight conformation in about half a second, ready to sense another blue photon. The molecular movie of the trans-to-cis isomerization of the photoactive center (a molecule of p-coumaric acid, PCA) of PYP, was “filmed” between 100fs and 3ps. The movie reveals that PYP remains in an excited state for about 600 fs upon light absorption before the chromophore atoms, still in trans configuration, begin to shift and continue over a 3ps time delay where the chromophore is in a cis configuration.
The Molecular movie of the trans/cis isomerization can be watched here: https://www.youtube.com/watch?v=b-CyE1f08Uk&t=3s
References:
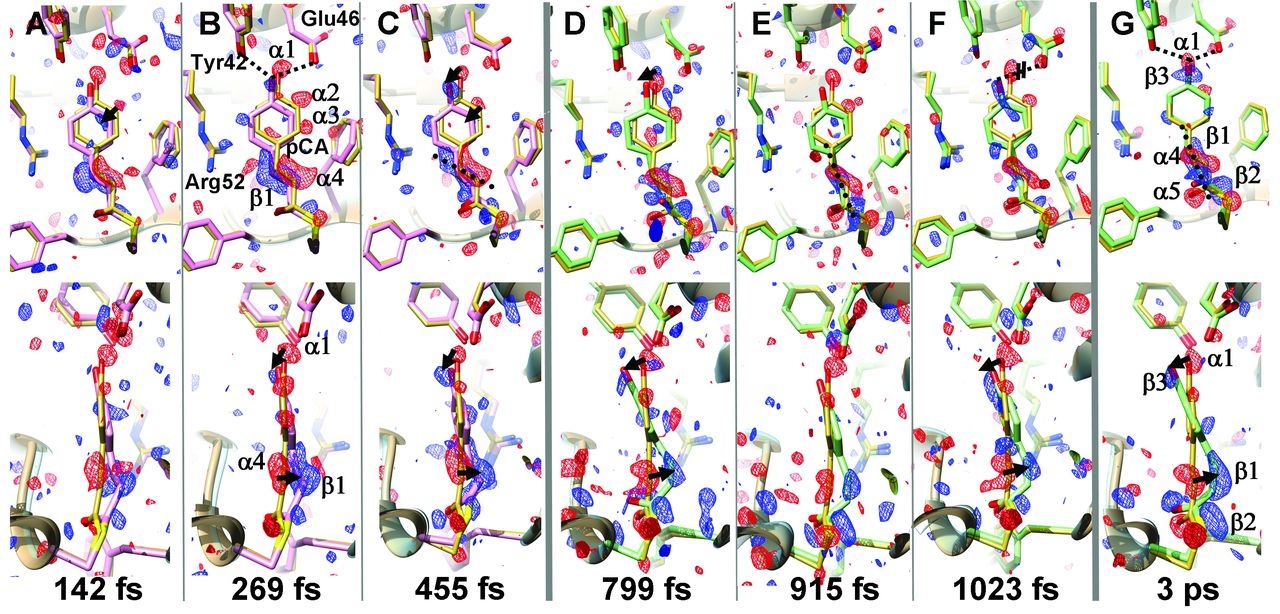
Ultrafast changes in the chromophore of PYP capture by TR-SFX. The PCA chromophore remains in trans configuration (purple structure) after 250 fs, and 3 ps after light excitation the configuration becomes cis (green structure). The yellow structure represents the dark state. Adapted from Pande et al. (2016).
4. Discovering one of the Secrets of Nature: The water Splitting Mechanism
Despite its crucial role in creating and sustaining life on Earth, photosynthesis is still not fully understood. One of its best kept secrets in nature is how photosystem II, a key protein complex in nature, harvests energy from sunlight and uses it to split water and produce the oxygen we breathe. A better understanding of how this process works could provide a blueprint for developing clean sources of renewable energy. Until recently, it had only been possible to measure fragments of this process at extremely low temperatures. With XFELs, scientists can now study and image steps of the process at its natural temperature towards obtaining the desired ‘molecular movie’ of photosystem II at work. The ultrabright and ultrafast X-ray pulses produced at XFELs are used to obtain maps of the molecular structure with atomic resolution and of how electrons flow in the oxygen-evolving complex of photosystem II. This allowed the researchers to narrow down the proposed mechanisms put forward by the research community over the years. Most recently, they were able to track the position of the oxygen atoms and the heavier metal atoms in the molecule, capturing all four metastable states of the process as well as some fleeting steps in between, providing unprecedented insight into the mysteries of the water splitting mechanism.
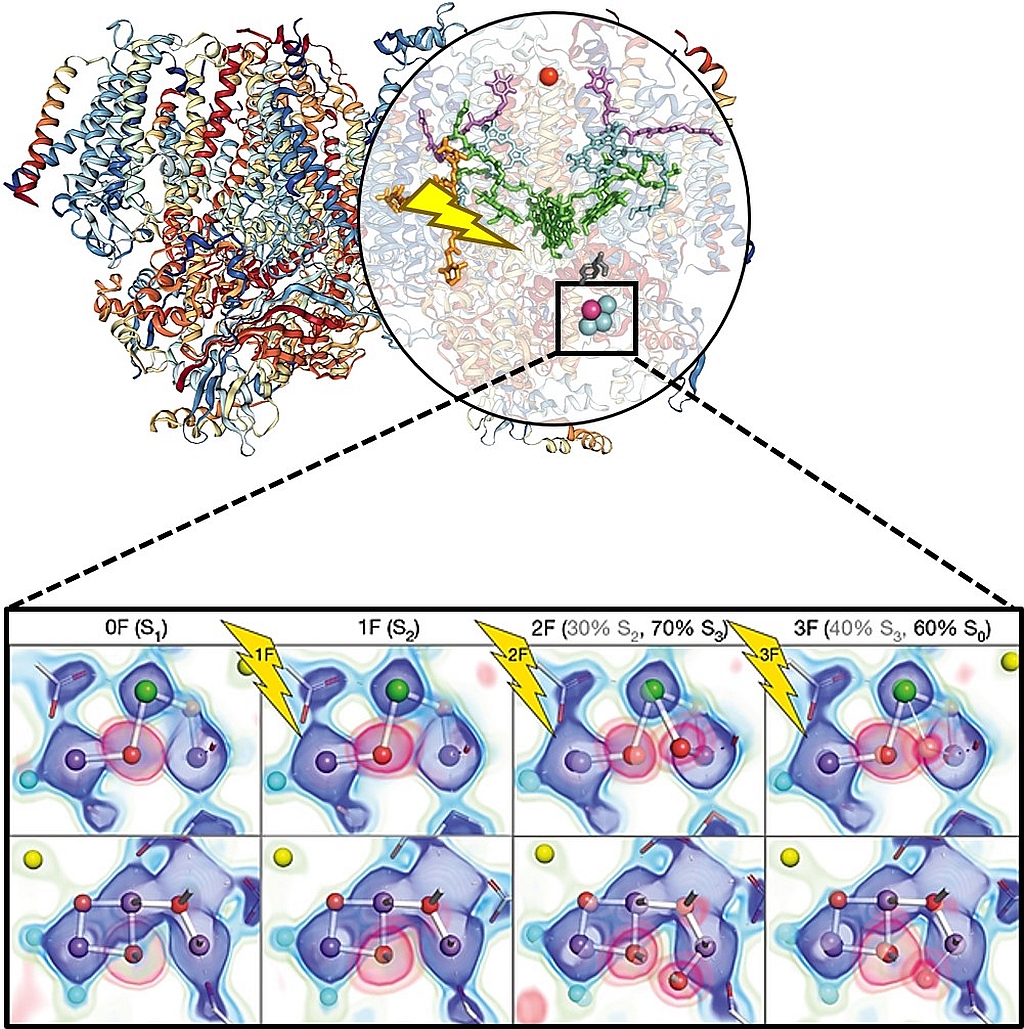
Figure shows the 3D model of photosystem II (left) and the stepwise changes occurring at the OEC during the oxygen-evolving Kok cycle (inset panel, adapted from Kern et al., 2018). Electron density and omit maps around OEC atoms are shown as the overlay of several contour levels for the two views of the OEC in the 0F-3F states. The contributions of the S states to each data set for the two-component analysis are indicated in parentheses.
5. Deciphering the Mechanism of a Biological Pump
Retinal is a chromophore bound to proteins called opsins, central to several important light-driven processes in people, animals, microbes, and algae, including human vision and some forms of photosynthesis, and the movie shows it changing shape in a trillionth of an eye blink. In the human eye, retinal begins in an cis configuration, which — upon capturing a photon of the correct wavelength — straightens out into a trans configuration. This configuration change pushes against an opsin protein in the retina, which triggers a chemical signaling cascade, which can result in perception of light or images by the human brain. The absorbance spectrum of the chromophore depends on its interactions with the opsin protein to which it is bound, so that different retinal-opsin complexes will absorb photons of different wavelengths (i.e., different colors of light). Retinal is so central to human vision that scientists have been studying it for nearly a century, steadily building a more detailed picture of how it works. In the past, scientists had to fill the gaps in their knowledge about retinal’s behavior by making inferences based on theory and computer simulations. Now, the first molecular movie of the instant when light hits retinal that’s widely used in nature for probing the environment and harvesting energy from light. The super-short pulses of XFELs allowed to collect data on the primary proton transfer events to identify where the atoms were in space and how that changed over time upon light excitation.
The molecular movie of the trans/cis isomerization of retinal: https://www.youtube.com/watch?v=c5n-OzkmUqc
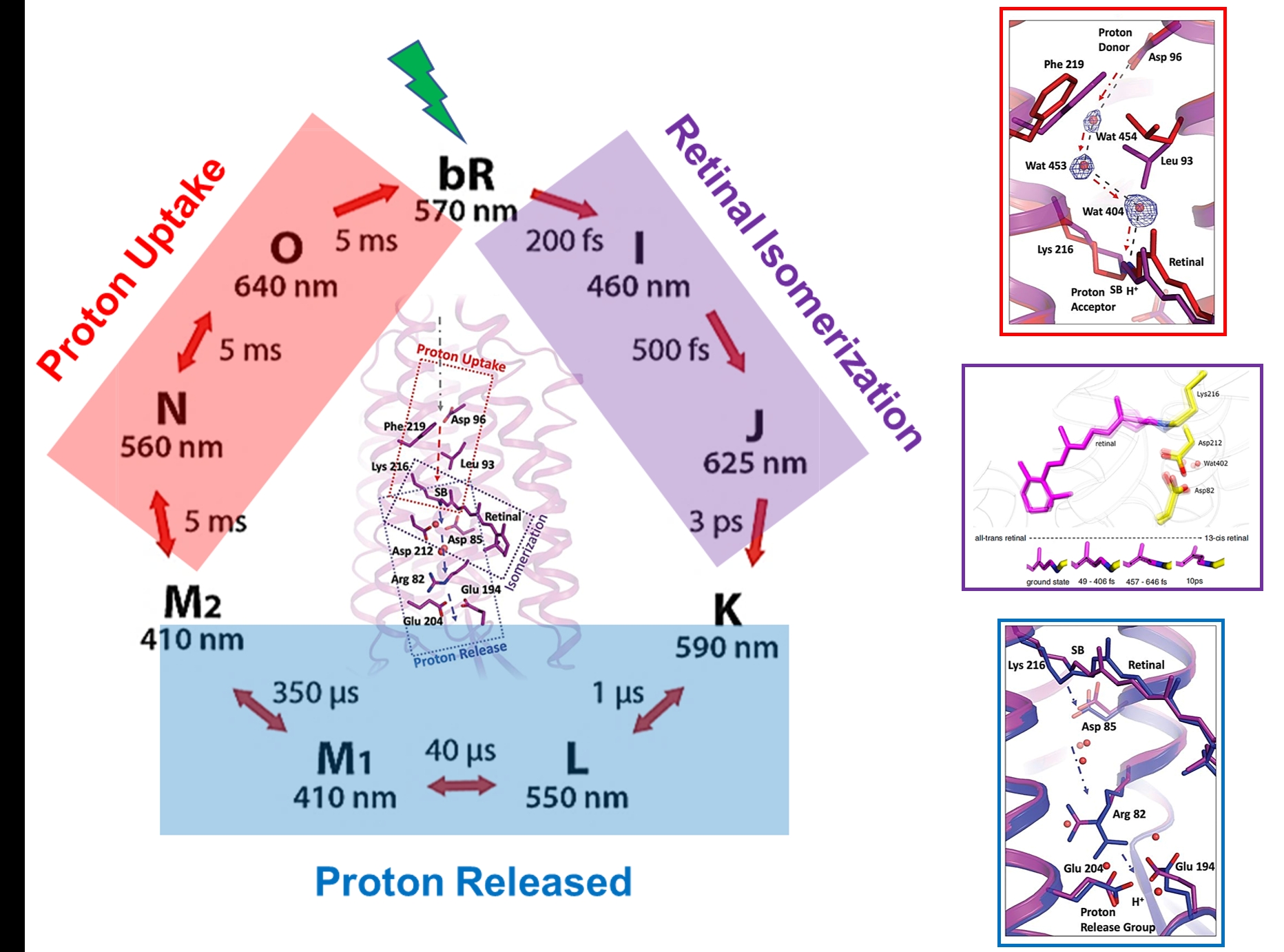
Bacteriorhodopsin (bR): The molecular movie of the photocycle of bR has been constructed at time delays from femtosecond to millisecond regimes. From these experiments the intermediates involved in the isomerization of the retinal molecule, the structural rearrangements undergone by bR, as well as the water molecule network acting as a proton wire, were all captured as a function of time.
6. Fighting Antibiotic Resistance by Watching Enzymatic Reactions in Real Time
Observing the catalytic cycle of an enzyme has been the goal of structural biologists since the first enzyme structure was solved. Because so many functions are dependent on enzymes, understanding their kinetics and dynamics has never been more important, unfortunately studying an enzyme while it is undergoing the catalytic cycle has always been difficult. The proof-of-concept of a mixing experiment at XFELs was done for watching, in “real time”, the inactivation of an antibiotic (Ceftriaxone, CEF) by the enzyme BlaC from Mycobacterium tuberculosis. Reaction intermediates were captured by collecting data sets at time points between 5ms to 2s upon mixing the protein crystals with CEF. From these experiments, it has been discovered that CEF binds BlaC shortly after mixing and that the nucleophilic attack by Serine70 and the subsequent opening of the β-lactam ring, occur 500ms later. Thus, watching the structure of enzymes attacking antibiotics can lead researchers to design more effective antibiotics to further hinder bacterial infections or even work on preventing antibiotic resistance.
The molecular movie of the binding of the antibiotic CEF can be watched here: https://www.youtube.com/watch?v=MbUefYBTObw
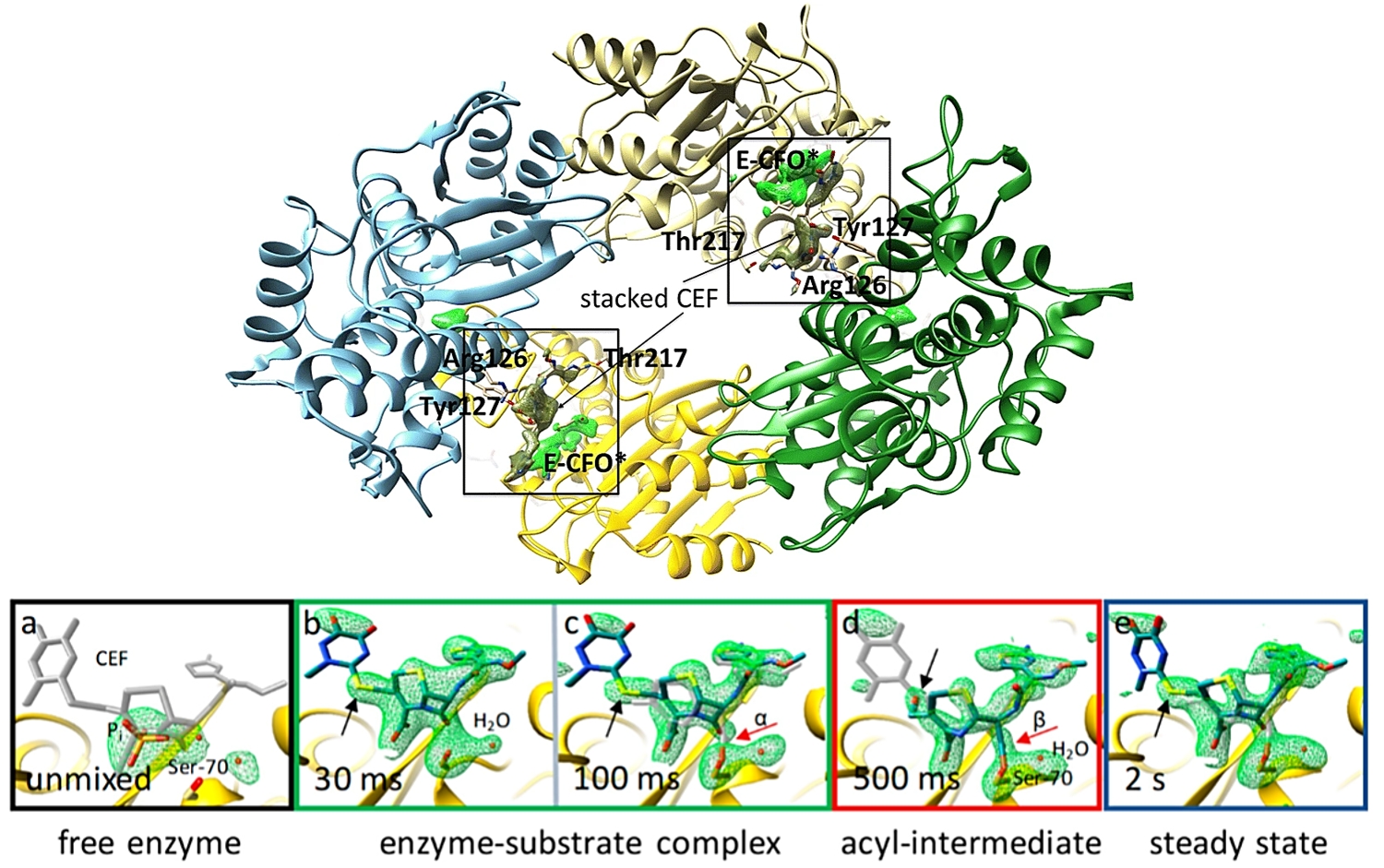
Top. Overview of BlaC at 500 ms after mixing with CEF. The omit maps (maps calculated after excluding atoms in question from the structure) are shown for the covalently bound intermediate E-CFO* in green. Electron density of an additional, stacked ceftriaxone molecule near the active site is shown in dark green. Figure adapted from Olmos et al., BMC Biol. (2018). Bottom. Simulated annealing omit maps of the active site structures (a–e) during the reaction at time points between 30 ms and 2 s upon mixing are shown. Figure adapted from Fromme et al., eLS (2020)
6. Watching Adenine Riboswitches in Action
Riboswitches are structural RNA elements that are generally located in the 5′ untranslated region of messenger RNA. During regulation of gene translation, ligand binding to the aptamer domain of a riboswitch leads to conformational changes that trigger a change in the ability of the mRNA to be translated by the ribosome. The crystallographic structures of the adenine riboswitch aptamer domain during the course of the reaction mechanism have been determined using mixed-and-inject serial crystallography at an XFEL with a 10s-time delay. The microcrystals of adenine riboswitch revealed a large change in the molecule’s shape with at least four transient states and illustrating how the structure of the molecule is linked to the way signals are transmitted. Also, analysis of the structures of intermediate and bound states reveals a 30° rotation of the riboswitch molecules in the crystals, leading to a change in the crystalline lattice. In addition, the binding of adenine induces large conformational changes that result in a polymorphic phase transition and lattice conversion from a monoclinic to an orthorhombic space group.
A molecular movie can be watched here: https://www.youtube.com/watch?v=MbUefYBTObw
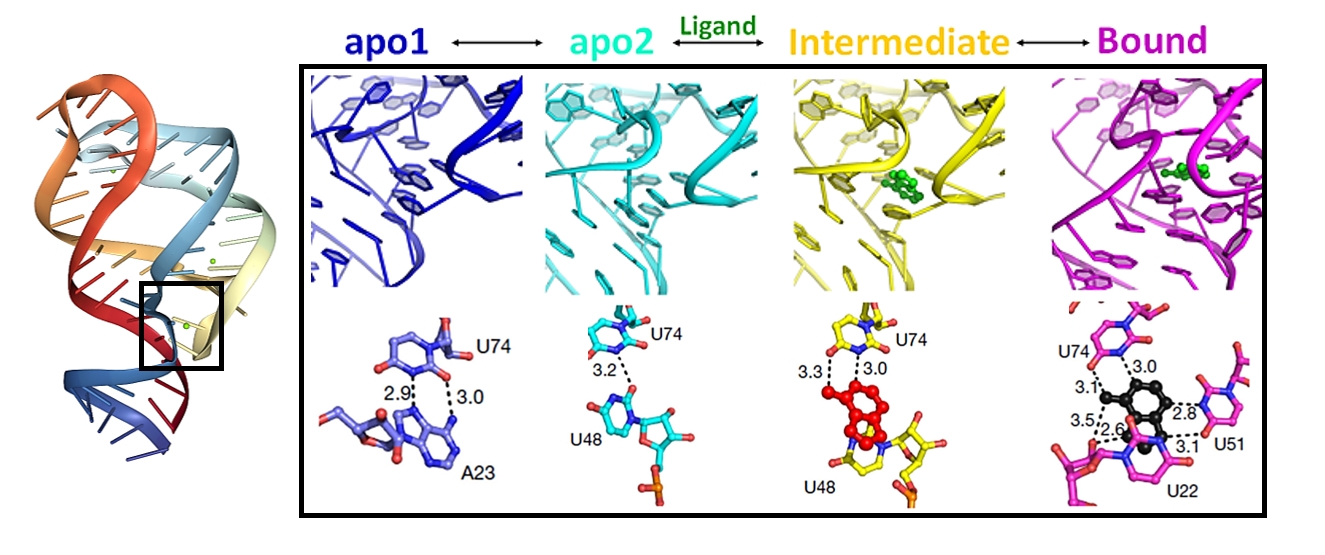
MISC experiment of a riboswitch RNA during its reaction with adenosine followed by a 10-s delay. A closer view of the active site of apo1, apo2, intermediate (IB) and ligand-bound states is illustrated in the box. Lower panels show the key residues in the ligand-binding pockets of all protein states.
